单载体表达系统Sigma-Aldrich 代谢组学
【简单介绍】
| 品牌 | Sigma-Aldrich | 货号 | 单载体表达系统 |
|---|---|---|---|
| 规格 | 糖酵解代谢 | 供货周期 | 一周 |
| 主要用途 | 脂肪酸/胆固醇代谢 |
继ZFN(Zinc Finger Nucleases)技术后,Merck在2013年推出新一代基因组编辑工具--CRISPR/Cas9,让研究人员以更快、更经济的方式实现基因组特定位点的编辑。
【详细说明】
基因组编辑工具-CRISPR/Cas9 | ||

继ZFN(Zinc Finger Nucleases)技术后,Merck在2013年推出新一代基因组编辑工具--CRISPR/Cas9,让研究人员以更快、更经济的方式实现基因组特定位点的编辑。凭借过去10年在基因组编辑领域的丰富经验积累以及专业的生物信息学平台,Merck已经成功设计出覆盖人类,小鼠和大鼠三个物种的所有基因的CRISPR/Cas9载体,并可以提供在线定制服务,以及完整的CRISPR实验workflow解决方案。此外,默克与Sanger Institute合作开发了人、小鼠全基因组CRISPR 文库,以帮助科学家实现基因功能的快速筛选、规模化的模型建立以及药物作用筛选等。
- 高效:优化的载体设计,大限度提高转染效率,简化筛选工作
- 特异:特殊的gRNA设计和双切口酶系统,大限度提高特异性
- 全面:产品齐全,可提供质粒、RNA、慢病毒载体、RNP等形式,涵盖人、大鼠、小鼠、植物等多个物种,更有Sanger Arrayed和Broad Pools全基因组文库以及重要通路的亚文库
- 掌控:强大的慢病毒全基因组文库可轻松进行高通量筛选,全面掌控人或小鼠的基因组
 |
| CRISPR/Cas9 基因编辑工具 |
| • Sanger Arrayed Lentiviral CRISPR Libraries • Lentiviral CRISPR Pools Libraries • CRISPR/Cas9单载体表达系统 • CRISPR Cas9-D10A双切口酶系统 • SygRNA® Cas9 RNP系统 • CRISPR/Cas9基因激活表达载体 • CRISPR/Cas9在植物中的应用 • Cas9蛋白 • CRISPR对照 (DNA and Virus) |
CRISPR/Cas Nuclease RNA-guided Genome Editing
Sigma-Aldrich is proud to offer its newest line of genome editing tools, Sigma CRISPRs, to the global research community. As the first company to commercially offer targeted genome editing technology nearly ten years ago, no one has more expertise in this field than Sigma-Aldrich. This experience is especially important when it comes to crafting genome editing tools that possess the critical requirements of having specific targeting and robust cutting activity. We provide both in our new Sigma CRISPR product line, and now we put our skill into your hands with a quick and simple web-based design platform. Sigma CRISPR products can be ordered directly through the link below or browse the CRISPR content on this page to learn more about the technology.
Order CRISPR Reagents
Sign up to receive the latest updates on CRISPR technology such as new journal articles, protocols, and product launches.
On this page:
What is CRISPR/Cas?
How does CRISPR/Cas work?
How Does CRISPR Cas9 Work?
What is CRISPR/Cas9?
All-in-one, ready-to-use Cas9 and guide RNA (gRNA) expression plasmids
CRISPR paired nickases
Purified RNA-only guide RNA
CRISPR Lentivirus
CRISPR positive and negative controls
What is CRISPR/Cas?
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. The discovery of the type II prokaryotic CRISPR “immune system” has allowed for the development for an RNA-guided genome editing tool that is simple, easy and quick to implement.
How does CRISPR/Cas work?
The CRISPR pathway was discovered in bacteria, where it functions much like an immune system against invading viruses and plasmid DNA. Short DNA sequences (spacers) from invading viruses are incorporated at CRISPR loci within the bacterial genome and serve as “memory” of previous infection. Re-infection triggers the complementary mature CRISPR RNA (crRNA) to find a matching sequence – which provides the CRISPR-associated (Cas) nuclease the specificity to form a double-strand break at specific “foreign” DNA sequences.
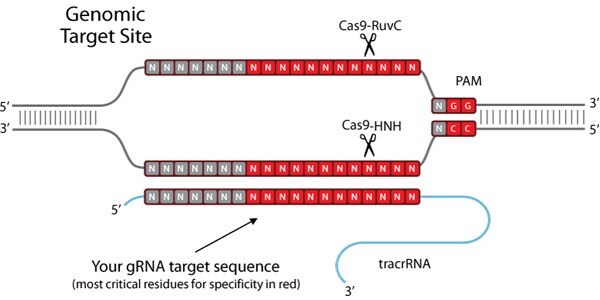
How Does CRISPR Cas9 Work?
To create such a tool, the endogenous CRISPR pathway was reduced to two principal components: the Cas9 nuclease and a guide RNA (gRNA)1-7. The guide RNA is a two component system consisting of the crRNA and tracrRNA. The crRNA targets the double stranded DNA to be cut, and has a short region of homology allowing it to bind the tracrRNA. The tracrRNA provides a stem loop structure which associates with Cas9 protein. The crRNA:tracrRNA duplex is referred to as the gRNA. The Cas9 nuclease and gRNA form a Cas9 ribonucleoprotein (RNP), which can bind and cut a specific DNA target in a whole genome context. In order to be cleaved by the RNP, a target must possess two specific sequences. First, the gRNA requires 17–21 bases of RNA-to-DNA homology, which is called the protospacer. Second, the Cas9 protein requires a short protospacer adjacent motif (PAM) in order to bind to the target DNA (see Figure 1, below right). If linking tracrRNA is present, and enough homology exists between the gRNA and the genomic target, the RNP cleaves both strands of the target DNA, creating a DSB at this precise location in the genome.

Figure 1. Schematic of a CRISPR/Cas-Targeted Double Strand Break.
While the functional CRISPR in the nucleus is in RNP form, the elements of CRISPR as a molecular tool are amenable to various methods of delivery. Early experiments succeeded in creating a chimeric single guide RNA, or sgRNA, that combines the crRNA and tracrRNA into a single RNA strand rather than the duplex found in nature. This sgRNA and Cas9 mRNA can be expressed from a single plasmid for either direct transfection or packaging into particles for lentiviral transduction. Alternatively, recombinant Cas9 protein can be combined with synthetically produced crRNA and tracrRNA to create an RNP for transfection or microinjection to embryos.
Start your genome editing experiment using our new gRNA design tool
Order SpCas9
Need help using Cas9 proteins?
Following targeted DSB formation, the cell typically uses one of two DNA repair pathways to survive: non-homologous end joining (NHEJ) or homology dependent repair (HDR). These repair mechanisms often make mistakes, resulting in mutagenesis at the target location either by disrupting a coding sequence to functionally inactivate or knockout a gene (NHEJ) or by adding new DNA sequence to knock-in specific sequence changes (HDR). In this way, the CRISPR/Cas9 system enables permanent, heritable modifications of chromosomal DNA. In addition, the CRISPR system can be used as a targeted delivery system when Cas9 endonuclease domains are inactivated and attached to other effector molecules to act at the gRNA-specified site in the genome. This expands the CRISPR system functions to gene activation and inhibition. Finally, the CRISPR system can be used for screening. All four primary uses of the CRISPR/Cas9 system—gene knockout, gene knock-in, gene activation or inhibition, and screening—are further discussed below.
Gene Knockout
CRISPR/Cas9 generates knockout cells or animals when co-expressed with a gRNA specific to the gene to be targeted. The purpose of gene knockout is to reveal gene function by disrupting its expression in the cell. A co-expressed, properly designed gRNA directs Cas9 to cleave a target sequence and generate a DSB in the gene of interest. Gene knockout can then be achieved in any one of three ways: 1) the cell repairs the break via NHEJ, leading to random insertions or deletions (“indels”) within the open reading frame (ORF) of the cleaved gene; 2) the cell repairs the break via HDR from a user-supplied template, inserting a specific disruptive sequence into the ORF; 3) a pair of gRNAs create two DSBs that flank an essential coding sequence, resulting in its excision.
NHEJ is the most active repair mechanism, but is error-prone and will often introduce a frameshift, causing a premature stop codon and/or causing the resulting transcript to be targeted for nonsense-mediated decay (NMD). Frameshift modifications can cause premature stop codons to be introduced into the transcript, preventing critical parts of the amino acid chain from being translated, resulting in an abnormally short, nonfunctional protein. Nonsense-mediated decay functions to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. It is theorized that this surveillance pathway is activated when exon-exon junction complexes—protein complexes that form on the pre-mRNA strand between exons during RNA splicing—are not properly removed by the ribosome after a transcript is spliced. When an exon-exon junction complex remains bound due to an upstream frameshift, the transcript is signaled for degradation and the functional protein is never translated. Both of these pathways are effective in disrupting gene function in a cell.
Gene Knock-in
CRISPR/Cas9 can also be used to introduce, or “knock in”, new DNA sequences. Common modifications include the introduction of a single nucleotide polymorphism (SNP), small tag, loxP, or a larger cassette such as a fluorescent protein. These modifications are made through a Cas9-induced DSB at a specific location, which significantly enhances the opportunity for targeted integration. Targeted integration (gene knock-in) occurs through HDR. In order to enable gene editing by HDR, a DNA ‘donor’ or repair template containing the desired sequence must be delivered to the cell, usually on a donor plasmid or oligonucleotide, along with the gRNA and Cas9. The efficiency of gene knock-in is generally lower than for knockout (<10% of modified alleles), but can be used to generate specific modifications ranging from a single nucleotide change to large insertions.
Gene Activation and Inhibition
In addition to functioning as a genome editing tool, CRISPR can also be used as a targeted delivery system for other functional proteins. A unique feature of Cas9 is its ability to bind target DNA independently of DNA cleavage, as these are two separate steps of Cas9’s mechanism. Wild-type Cas9 has two nuclease domains: RuvC and HNH. To achieve binding without cleavage, both nuclease domains are rendered inactive by inducing point mutations (D10A and H840A in SpCas9), resulting in a nuclease dead Cas9 (dCas9). When combined with gRNAs targeting transcriptional start sites, it was discovered that dCas9 alone was sufficient to decrease or inhibit transcription by blocking transcription initiation. Following this development, scientists began experimenting with transcriptional repressors and activators tethered to dCas9. The KRAB transcriptional repressor domain has been fused to dCas9 as a mode of effector domain-mediated transcriptional silencing. dCas9 has also been fused to transcriptional activators and transcriptional effector proteins. dCas9 for activation is a widely popular area of study and includes systems such as, dCas9-VP64, dCas9_SunTag, dCas9-SAM, dCas9-RNA Scaffold, and dCas9-VPR. At MilliporeSigma, we have fused dCas9 to the catalytic histone acetyltransferase (HAT) core domain of the human E1A-associated protein P300. When combined with gRNAs targeting transcriptional start sites and enhancer regions, dCas9-P300 directs histone acetylation, releasing DNA from its heterochromatic state to upregulate gene expression via endogenous cellular machinery in the native chromosomal context. The dCas9-P300 histone acetylation approach represents a distinct mechanism of action relative to dCas9-VP64 or other similar gene activation motifs.
DCAS9P300
Screening
Screens enable a quick survey of large numbers of candidate genes or genomic sites for involvement in your pathway or phenotype of interest. Improvements in pooled oligo manufacturing and big data handling, combined with the efficiency of lentiviral delivery, allow researchers to consider thousands of candidate genes at once, either as a library or a more focused panel. We define libraries as whole genome collections of CRISPR clones; panels are smaller gene subsets. Generally, panels and libraries may be assembled in two formats: pooled (thousands of gRNA in one tube) or arrayed (one gRNA per well in 96-well plates). Both approaches provide rapid answers to larger questions, offering significant advantages over past methods that required a time- and labor-intensive process of interrogating candidate genes one at a time.
MilliporeSigma offers a variety of screening solutions, including the pooled GeCKO libraries from the Broad and the Sanger Arrayed Whole Genome Lentiviral CRISPR Library to perform large gene knockout screens(8). And, coming soon are the human and mouse CRISPR/Cas9 Synergistic Activation Mediator (SAM) pooled libraries for genome-wide screening of transcriptional activation phenotypes!
SANGER LIBRARY
What is CRISPR/Cas9?
CRISPR/Cas9 systems were discovered in 1993, and shown to have evolved within bacterial and archaeal organisms as a defense against viruses and foreign plasmids. The function of the native CRISPR pathway is to target nucleases to invasive DNA, creating a potentially lethal double-strand break (DSB). Shortly after the function of this pathway was recognized, the type II CRISPR/Cas9 system from the bacterium Streptococcus pyogenes (SpCas9) was repurposed to create a simple, yet powerful molecular tool that could be programmed to target nucleases to a specific genomic sequence.
All-in-one, ready-to-use Cas9 and guide RNA (gRNA) expression plasmids
- A codon optimized Cas9 protein and a gRNA are expressed from a single vector and provided as ready-
to-use, transfection-grade DNA. - Unique CRISPR sites pre-designed to minimize off-targeting are available for the coding regions of the human, mouse, and rat genomes. Custom designs for any other regions or species are always available upon request (see Custom CRISPR request form).
- GFP or RFP is fused to the C-terminus of Cas9 with a 2A peptide, enabling tracking of transfection efficiency and enrichment of genome editing activity through FAC sorting.


CRISPR paired nickases
- Sigma′s CRISPR paired nickases further minimize off-target double-stranded breaks by requiring the independent binding of two separate gRNAs to a localized genomic region.
- In the presence of the Cas9-D10A nicking nuclease, the two gRNAs induce single-stranded breaks on opposite strands of the DNA, creating a functional double-stranded break.
- Pre-designed, unique CRISPR paired nickases are available for the coding regions of the human, mouse, and rat genomes (Paired Nickases Predesigns). Designs for any other regions or species are available as a custom request (see Custom CRISPR request form).
- When ordering Sigma′s CRISPR paired nickases for a specific target, researchers will receive two separate ready-to-use, transfection-grade plasmids expressing gRNA from the human U6 promoter. The Cas9-D10A nicking nuclease must be purchased separately as either plasmid or mRNA.


Paired Cas9 Nickases - For optimal Cas9-D10A paired nickase functionality, guide RNAs should be designed in a 5’-to-5’ orientation with PAM spacing between 30 and 150 bp.

Purified RNA-only guide RNA
- Ready-to-use gRNAs in a purified RNA format suitable for microinjection or cell culture.
- Designed using the same stringent rules as our Sigma CRISPRs and CRISPR paired nickase plasmids.
- The CRISPR RNA format is ideal for researchers creating transgenic animal models or for those concerned about genomic plasmid integration.
- The RNA format avoids the need for a specific promoter, allowing for expression in most known cell types and organisms.
- Wild type Cas9 (CAS9P) or Cas9-D10A nicking (CAS9D10AP) nuclease must be purchased separately as either plasmid or mRNA (CAS9MRNA, CAS9D10AMRNA).
- Donor Designs available.

CRISPR Lentivirus
- Sigma CRISPR Lentivirus for high efficiency knockout screens
- The lenti format allows for efficient chromosomal integration of CRISPR components.
- Transduce cells using Sigma’s unique single lentiviral CRISPR format, which features a Cas9 ORF flanked by puro and GFP elements, providing multiple options for monitoring stable cell populations.
- Enables genome editing even in difficult to transfect cells.
- Choose from our pre-designed CRISPR sites or submit custom targets

Sigma Lentiviral CRISPR: (A) Vector map of the all-in-one lenti CRISPR vector. (B) GFP signal post-transduction of HEK293 cells with particles made via pLV-U6g-EPCG. (C) HeLa cells were transduced with lentivirus (pLV-U6g-EPCG) followed by selection on puromycin and 6TG treatment to enrich for HPRT1 knockouts. Insertions and deletions were detected at the gRNA target site via mismatch assay (CEL-I).

CRISPR positive and negative controls
- A tested and validated positive control for the human EMX1 gene provides a basis of reference and allows researchers to perform side-by-side assessments of the efficiency of their genome editing experiments.
- Ready-to-use human EMX1 positive controls for both single gRNAs (CRISPR01-1SET) and CRISPR paired nickases (CRISPR02-1SET) are available.
- Each human EMX1 positive control comes with an aliquot of either the wild type Cas9 (CRISPR01-1SET) or nickase Cas9-D10A nuclease (CRISPR02-1SET).
- CRISPR Universal negative controls (CRISPR06-1EA, CRISPR07-1EA, CRISPR08-1EA) are all-in-one, ready-to-use, transfection-grade plasmids expressing the wild type Cas9 nuclease under the CMV promoter, along with and a gRNA sequence designed to target no known human, mouse, or rat gene.

CEL I image of CRISPR01 positive control (lane labeled EMX1s4+Cas9) and CRISPR02 positive control (lane labeled EMX1s4, EMX1as4+D10A)
CRISPR/Cas9 Vector
For additional information and to customize and purchase CRISPR click Order CRISPR below
Materials
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
相关产品
- RestrictionEnzymSigma-Aldrich 代谢组学克隆和表达
- SnapFast VectorsSigma-Aldrich 代谢组学克隆和表达
- Cloning EnzymesSigma-Aldrich 代谢组学克隆和表达
- CRISPR对照Sigma-Aldrich 代谢组学
- 在植物中的应用Sigma-Aldrich 代谢组学
- Cas9蛋白Sigma-Aldrich 代谢组学
- 基因激活表达载体Sigma-Aldrich 代谢组学
- SygRNA® Cas9 RNPSigma-Aldrich 代谢组学
- 双切口酶系统Sigma-Aldrich 代谢组学
- 单载体表达系统Sigma-Aldrich 代谢组学
- Lentiviral CRISPSigma-Aldrich 代谢组学
- 基因组编辑工具Sigma-Aldrich 代谢组学
- 酶活性检测Sigma-Aldrich 代谢组学
- 脂肪酸/胆固醇Sigma-Aldrich 代谢组学
- 糖酵解代谢Sigma-Aldrich 代谢组学
- 氨基酸代谢Sigma-Aldrich 代谢组学
请输入产品关键字:
邮编:200437
联系人:郑经理
电话:13816982406,021-65010900
传真:021-65010900
手机:13816982406
留言:发送留言
网址:
商铺:https://www.chem17.com/st215208/


 QQ交谈
QQ交谈
