详细介绍
高通量单分子力谱仪系统
德国AFS单分子原子力谱仪 (Single Molecule Atomic Force Spectrometer)
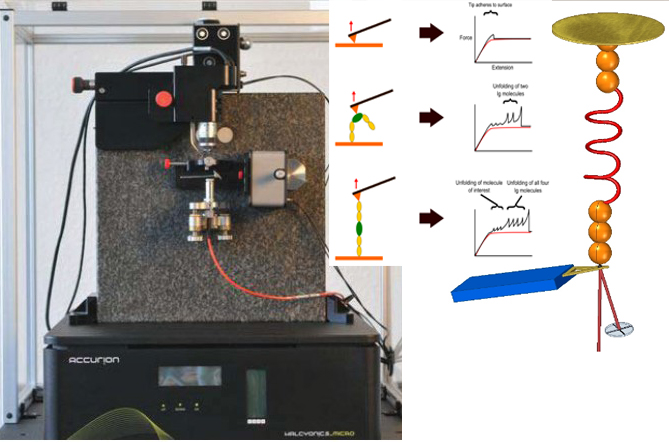
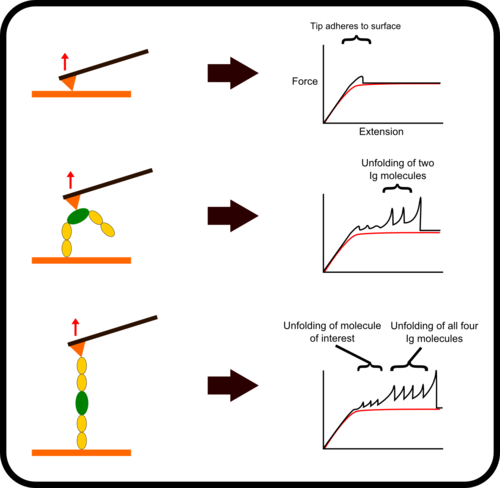

新型,高线性原子力光谱仪,用于蛋白质动力学的定量测量
该装置可对单个分子进行高通量分析,阐明诸如解折叠速率和保持其构象的键强度等特征。根据要确定的分子的性质,此设备可以与多种协议一起使用:力扩展,力钳,力斜坡和各种重新折叠协议。
- 力钳和力-延伸
- 亚纳米分辨率
- 亚毫秒级时间分辨率
- 蛋白质折叠和展开
- 键裂解和形成
- 全自动操作
- 强大的分析软件
- 简单的用户界面
- 高通量

原子力光谱仪(AFS)是用于在校准的机械负载下单个蛋白研究的仪器.
AFS用于了解机械力在整个生物光谱,影响蛋白质的动力学和化学性质。
AFS允许拾取和机械操纵单个重组蛋白。 该系统全自动,可以连续运行数天,无需人看管。
操作软件中包含的例程明确识别所研究蛋白质的机械指纹,并因此能够
自动识别和存储数据。在为期一天的实验结束后,可以收集多个一百种单蛋白痕迹。
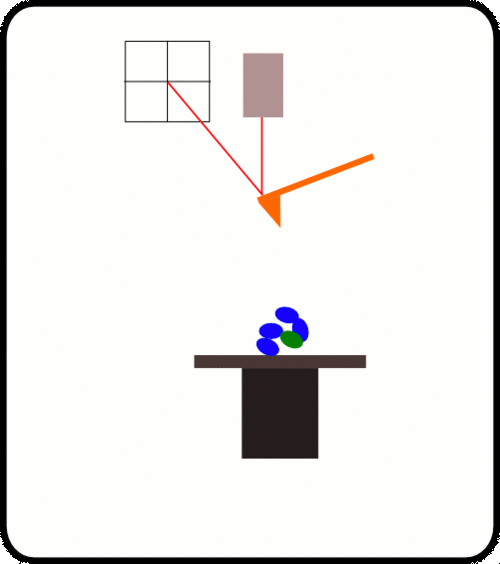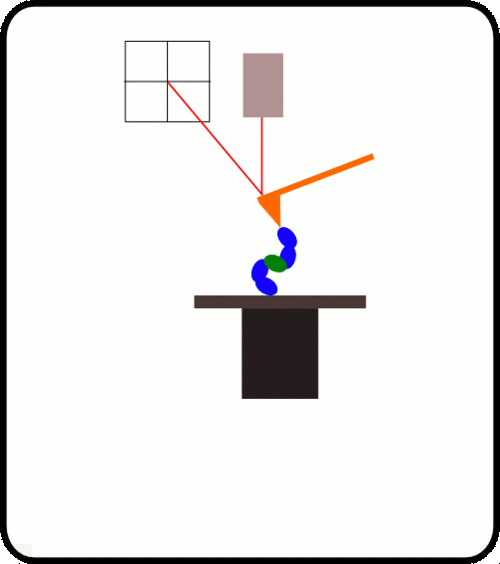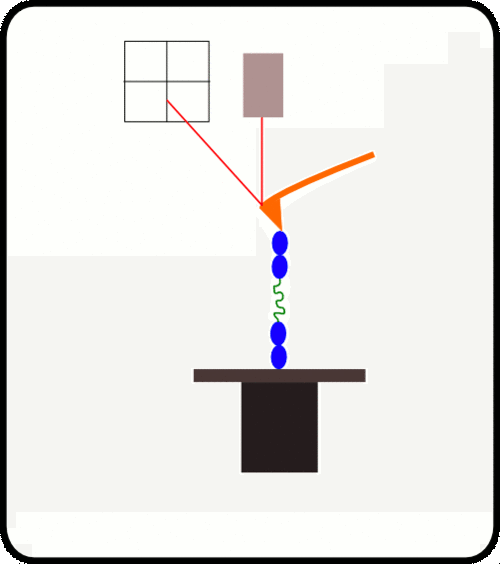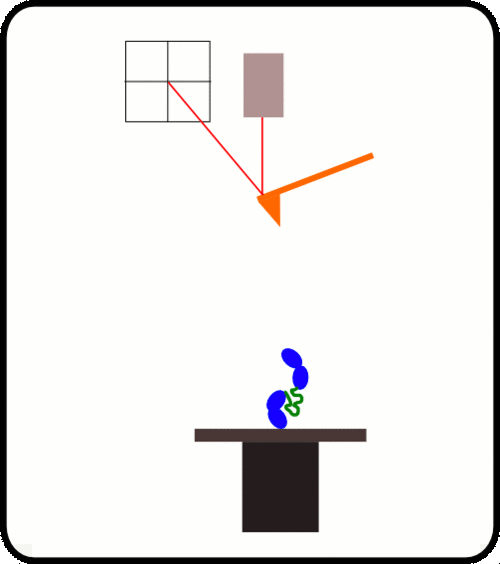
用于AFS测量的蛋白质附着的标准方法是使用硫醇化学方法,该方法是通过在覆盖有金的玻璃盖玻片上将在末端含有半胱氨酸锚定分子的重组蛋白分层来获得的。 zui有效的方法是结合使用HaloTag(Promega)技术和硫醇化学方法,该方法可以很容易地将共价连接的蛋白质传递给两者。 镀金的悬臂和Halo-配体的玻璃盖玻片。 共价连接的蛋白质可以长时间进行机械操作。
优选的蛋白质样品被安排为串联模块蛋白质。 当此类多蛋白通过AFS铺展时,其作用力是*的机械指纹,可将它们与困扰单分子研究的更常见的非特异性事件明确区分开。
This unique Atomic-Force-Spectroscope (AFS) , based on the Atomic Force Microscope Technology (AFM) was specially designed for studying single proteins placed under a calibrated mechanical load.
The upside down design, where the cantilever, laser and sensor is fixed while the substrate with attached protein is moved by a ultra-precise linear Piezo, enables us to get a feedback response time better than 1ms and a position accuracy better than 1nm.
The software is based on IGOR and enables the operator to run automatic experiments over the day without putting hands on.
The AFS can be operated in two modes:
- force extension (constant velocity)
- force clamp (constant force)
In force clamp mode the operator can design his own force protocol to measure protein folding, unfolding and protein chemical reactions.
The software includes routines that unambiguously identify the mechanical fingerprints of the protein being studied, and thus is able to recognize and store data automatically.
A tuneable Proportional-Integral-Differential system (PID) enables the operator to adjust the parameter for the Piezo, cantilever and probe to get the best feedback time for each individual experiment.
The AFS comes as a complete workstation, incl. vibration damping table, PC Monitor and software.
For further information please visit the web page of Professor Julio Fernandez, in which collaboration this system was designed.
We use a purpose built single-molecule force spectroscope (SMFS) developed by the Fernandez lab at Columbia University and built by Luigs Neumann.[1] This device allows for high throughput analysis of single molecules, elucidating characteristics such as the rate of unfolding and the strength of the bonds holding its conformation. This device can be used with a range of protocols depending on the property of the molecule to be determined: force-extension, force-clamp, force-ramp, and various refolding protocols.
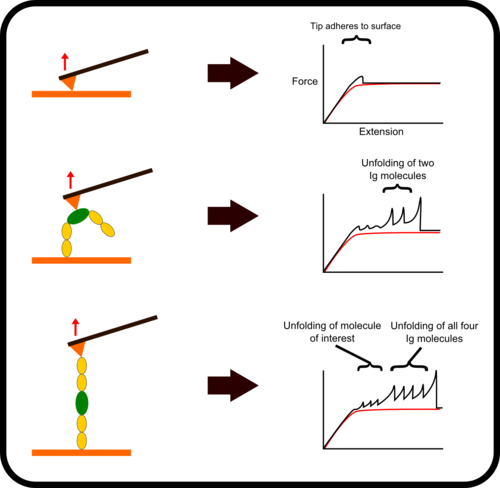
Chimera Fingerprinting
One of the main difficulties with high-throughput SMFS is that a great proportion of the data produced is not relevant for study as it does not the display the molecule under investigation. Non-specific binding of the cantilever tip at various points along the molecule leads to incomplete traces. One method to overcome this is to add specific immunoglobulin (Ig) domains at either end of the molecule being studied; these Ig domains unfold with very specific patterns which, if present in a trace, indicate the molecule has been correctly stretched. Thus thousands of force-extension curves can be sorted rapidly by requiring them to have the characteristic ‘saw-tooth’ pattern that indicative of the unfolding of an Ig domain.
Surface Attachment
In addition to using flanking molecules to improve the detection of correct stretching traces, surface chemistry can be used to covalently bind the end of the protein construct to surface or the tip. This increases the likelihood that a molecule will be stretched along its full length and thus increases the efficiency of the experiment. The most common way to do this is to prepare the surface such that it contains a particular ligand for an enzyme. This enzyme is then attached at the end of the protein construct, similarly to the fingerprint Ig domains. As such when the protein is deposited on the surface, the enzyme reacts with the functionalized surface to produce a strong covalent attachment between the surface and the protein construct.[2]
[1] Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy
Ionel Popa Pallav Kosuri Jorge Alegre-Cebollada Sergi Garcia-Manyes Julio M. Fernandez
Nature Protocols June, 2013
[2] Nanomechanics of HaloTag Tethers
Ionel Popa Ronen Berkovich Jorge Alegre-Cebollada Carmen L. Badilla Jaime Andrés Rivas-Pardo Yukinori Taniguchi Masaru Kawakami Julio M. Fernandez
Journal of the American Chemical Society August, 2013
Animation of AFS unfolding of a talin-Ig construct
 化工仪器网
化工仪器网






 化工仪器网
化工仪器网